Support Vector Machines (SVMs)¶
Support Vector Machines from [LIBSVM]¶
- class mlpy.LibSvm(svm_type='c_svc', kernel_type='linear', degree=3, gamma=0.001, coef0=0, C=1, nu=0.5, eps=0.001, p=0.1, cache_size=100, shrinking=True, probability=False, weight={})¶
LibSvm.
Parameters : - svm_type : string
SVM type, can be one of: ‘c_svc’, ‘nu_svc’, ‘one_class’, ‘epsilon_svr’, ‘nu_svr’
- kernel_type : string
kernel type, can be one of: ‘linear’ (uT*v), ‘poly’ ((gamma*uT*v + coef0)^degree), ‘rbf’ (exp(-gamma*|u-v|^2)), ‘sigmoid’ (tanh(gamma*uT*v + coef0))
- degree : int (for ‘poly’ kernel_type)
degree in kernel
- gamma : float (for ‘poly’, ‘rbf’, ‘sigmoid’ kernel_type)
gamma in kernel (e.g. 1 / number of features)
- coef0 : float (for ‘poly’, ‘sigmoid’ kernel_type)
coef0 in kernel
- C : float (for ‘c_svc’, ‘epsilon_svr’, ‘nu_svr’)
cost of constraints violation
- nu : float (for ‘nu_svc’, ‘one_class’, ‘nu_svr’)
nu parameter
- eps : float
stopping criterion, usually 0.00001 in nu-SVC, 0.001 in others
- p : float (for ‘epsilon_svr’)
p is the epsilon in epsilon-insensitive loss function of epsilon-SVM regression
- cache_size : float [MB]
size of the kernel cache, specified in megabytes
- shrinking : bool
use the shrinking heuristics
- probability : bool
predict probability estimates
- weight : dict
changes the penalty for some classes (if the weight for a class is not changed, it is set to 1). For example, to change penalty for classes 1 and 2 to 0.5 and 0.8 respectively set weight={1:0.5, 2:0.8}
- LibSvm.learn(x, y)¶
Constructs the model. For classification, y is an integer indicating the class label (multi-class is supported). For regression, y is the target value which can be any real number. For one-class SVM, it’s not used so can be any number.
Parameters : - x : 2d array_like object
training data (N, P)
- y : 1d array_like object
target values (N)
- LibSvm.pred(t)¶
Does classification or regression on test vector(s) t.
Parameters : - t : 1d (one sample) or 2d array_like object
test data ([M,] P)
Returns : - p : for a classification model, the predicted class(es) for t is
returned. For a regression model, the function value(s) of t calculated using the model is returned. For an one-class model, +1 or -1 is returned.
- LibSvm.pred_probability(t)¶
Returns C (number of classes) probability estimates. For a ‘c_svc’ and ‘nu_svc’ classification models with probability information, this method computes ‘number of classes’ probability estimates.
Parameters : - t : 1d (one sample) or 2d array_like object
test data ([M,] P)
Returns : - probability estimates : 1d (C) or 2d numpy array (M,C)
probability estimates for each observation.
- LibSvm.pred_values(t)¶
Returns D decision values. For a classification model with C classes, this method returns D=C*(C-1)/2 decision values for each test sample. The order is label[0] vs. label[1], ..., label[0] vs. label[C-1], label[1] vs. label[2], ..., label[C-2] vs. label[C-1], where label can be obtained from the method labels().
For a one-class model, this method returns D=1 decision value for each test sample.
For a regression model, this method returns the predicted value as in pred()
Parameters : - t : 1d (one sample) or 2d array_like object
test data ([M,] P)
Returns : - decision values : 1d (D) or 2d numpy array (M,D)
decision values for each observation.
- LibSvm.labels()¶
For a classification model, this method outputs the name of labels. For regression and one-class models, this method returns None.
- LibSvm.nclasses()¶
Get the number of classes. = 2 in regression and in one class SVM
- LibSvm.nsv()¶
Get the total number of support vectors.
- LibSvm.label_nsv()¶
Return a dictionary containing the number of support vectors for each class (for classification).
- static LibSvm.load_model(filename)¶
Loads model from file. Returns a LibSvm object with the learn() method disabled.
- LibSvm.save_model(filename)¶
Saves model to a file.
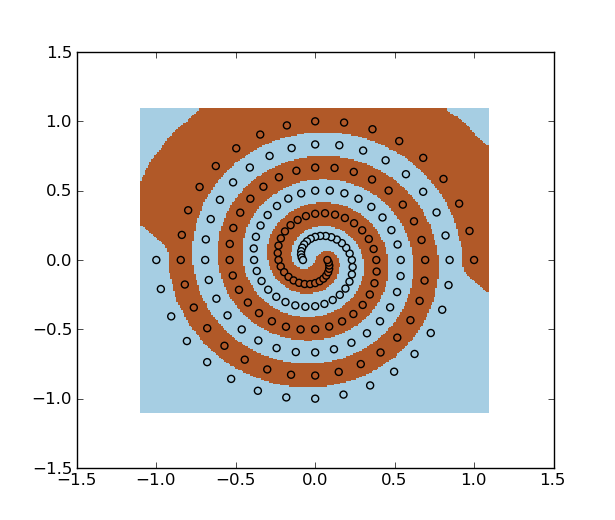
Example on spiral dataset:
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> import mlpy
>>> f = np.loadtxt("spiral.data")
>>> x, y = f[:, :2], f[:, 2]
>>> svm = mlpy.LibSvm(svm_type='c_svc', kernel_type='rbf', gamma=100)
>>> svm.learn(x, y)
>>> xmin, xmax = x[:,0].min()-0.1, x[:,0].max()+0.1
>>> ymin, ymax = x[:,1].min()-0.1, x[:,1].max()+0.1
>>> xx, yy = np.meshgrid(np.arange(xmin, xmax, 0.01), np.arange(ymin, ymax, 0.01))
>>> xnew = np.c_[xx.ravel(), yy.ravel()]
>>> ynew = svm.pred(xnew).reshape(xx.shape)
>>> fig = plt.figure(1)
>>> plt.set_cmap(plt.cm.Paired)
>>> plt.pcolormesh(xx, yy, ynew)
>>> plt.scatter(x[:,0], x[:,1], c=y)
>>> plt.show()
| [LIBSVM] | Chih-Chung Chang and Chih-Jen Lin. LIBSVM: a library for support vector machines. 2001. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm |
| [Cristianini] | N Cristianini and J Shawe-Taylor. An introduction to support vector machines. Cambridge University Press. |
| [Vapnik95] | V Vapnik. The Nature of Statistical Learning Theory. Springer-Verlag, 1995. |
Kernel Adatron¶
- class mlpy.KernelAdatron(C=1000, maxsteps=1000, eps=0.01)¶
Kernel Adatron algorithm without-bias-term (binary classifier).
The algoritm handles a version of the 1-norm soft margin support vector machine. If C is very high the algoritm handles a version of the hard margin SVM.
Use positive definite kernels (such as Gaussian and Polynomial kernels)
Parameters : - C : float
upper bound on the value of alpha
- maxsteps : integer (> 0)
maximum number of steps
- eps : float (>=0)
the algoritm stops when abs(1 - margin) < eps
- KernelAdatron.learn(K, y)¶
Learn.
- Parameters:
- K: 2d array_like object (N, N)
- precomputed kernel matrix
- y : 1d array_like object (N)
- target values
- KernelAdatron.pred(Kt)¶
Compute the predicted class.
Parameters : - Kt : 1d or 2d array_like object ([M], N)
test kernel matrix. Precomputed inner products (in feature space) between M testing and N training points.
Returns : - p : integer or 1d numpy array
predicted class
- KernelAdatron.margin()¶
Return the margin.
- KernelAdatron.steps()¶
Return the number of steps performed.
- KernelAdatron.alpha()¶
Return alpha
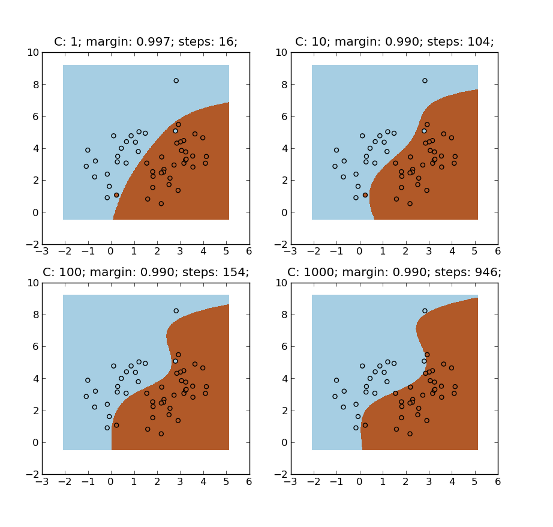
Example:
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> import mlpy
>>> np.random.seed(0)
>>> mean1, cov1, n1 = [1, 4.5], [[1,1],[1,2]], 20 # 20 samples of class 1
>>> x1 = np.random.multivariate_normal(mean1, cov1, n1)
>>> y1 = np.ones(n1, dtype=np.int)
>>> mean2, cov2, n2 = [2.5, 2.5], [[1,1],[1,2]], 30 # 30 samples of class 2
>>> x2 = np.random.multivariate_normal(mean2, cov2, n2)
>>> y2 = 2 * np.ones(n2, dtype=np.int)
>>> x = np.concatenate((x1, x2), axis=0) # concatenate the samples
>>> y = np.concatenate((y1, y2))
>>> K = mlpy.kernel_gaussian(x, x, sigma=2) # kernel matrix
>>> xmin, xmax = x[:,0].min()-1, x[:,0].max()+1
>>> ymin, ymax = x[:,1].min()-1, x[:,1].max()+1
>>> xx, yy = np.meshgrid(np.arange(xmin, xmax, 0.02), np.arange(ymin, ymax, 0.02))
>>> xt = np.c_[xx.ravel(), yy.ravel()] # test points
>>> Kt = mlpy.kernel_gaussian(xt, x, sigma=2) # test kernel matrix
>>> fig = plt.figure(1)
>>> cmap = plt.set_cmap(plt.cm.Paired)
>>> for i, c in enumerate([1, 10, 100, 1000]):
... ka = mlpy.KernelAdatron(C=c)
... ax = plt.subplot(2, 2, i+1)
... ka.learn(K, y)
... ytest = ka.pred(Kt).reshape(xx.shape)
... title = ax.set_title('C: %s; margin: %.3f; steps: %s;' % (c, ka.margin(), ka.steps()))
... plot1 = plt.pcolormesh(xx, yy, ytest)
... plot2 = plt.scatter(x[:,0], x[:,1], c=y)
>>> plt.show()
| [Friess] | Friess, Cristianini, Campbell. The Kernel-Adatron Algorithm: a Fast and Simple Learning Procedure for Support Vector Machines. |
| [Kecman03] | Kecman, Vogt, Huang. On the Equality of Kernel AdaTron and Sequential Minimal Optimization in Classification and Regression Tasks and Alike Algorithms for Kernel Machines. ESANN‘2003 proceedings - European Symposium on Artificial Neural Networks, ISBN 2-930307-03-X, pp. 215-222. |