Canberra Distances and Stability Indicator of Ranked Lists¶
Canberra distance¶
- mlpy.canberra(x, y)¶
Returns the Canberra distance between two P-vectors x and y: sum_i(abs(x_i - y_i) / (abs(x_i) + abs(y_i))).
Canberra Distance with Location Parameter¶
See [Jurman08].
- mlpy.canberra_location(x, y, k=None)¶
Returns the Canberra distance between two position lists, x and y. A position list of length P contains the position (from 0 to P-1) of P elements. k is the location parameter, if k=None will be set to P.
- The function computes:
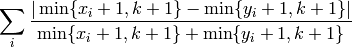
- mlpy.canberra_location_expected(p, k=None)¶
Returns the expected value of the Canberra location distance, where p is the number of elements and k is the number of positions to consider.
Canberra Stability Indicator¶
See [Jurman08].
- mlpy.canberra_stability(x, k=None)¶
Returns the Canberra stability indicator between N position lists, where x is an (N, P) matrix. A position list of length P contains the position (from 0 to P-1) of P elements. k is the location parameter, if k=None will be set to P. The lower the indicator value, the higher the stability of the lists.
The stability is computed by the mean distance of all the (N(N-1))/2 non trivial values of the distance matrix (computed by canberra_location()) scaled by the expected (average) value of the Canberra metric.
Example:
>>> import numpy as np >>> import mlpy >>> x = np.array([[2,4,1,3,0], [3,4,1,2,0], [2,4,3,0,1]]) # 3 position lists >>> mlpy.canberra_stability(x, 3) # stability indicator 0.74862979571499755
| [Jurman08] | (1, 2) Jurman, Riccadonna, Visintainer and Furlanello. lgebraic stability indicators for ranked lists in molecular profiling. Bioinformatics Vol. 24 no. 2 2008, pages 258–264. |